CrysAlisPro Tip
Proffitloop
What does it do?
Proffitloop is a feature within CrysAlisPro that allows one to run automated multiple data reductions successively with different PROFFIT settings on a given dataset.
Why should I use it?
In order to get the best result from each single diffraction dataset, one can try different parameter combinations during data reduction manually, although this can be quite time-consuming. Proffitloop allows for automated batch processing of data with certain parameters switched on and off for successive runs. Proffitloop should especially be used for problematic or difficult single component datasets; e.g., with varying background, high mosaic crystals or fluorescence. Twinned datasets are currently not supported by Proffitloop.
How do I use it?
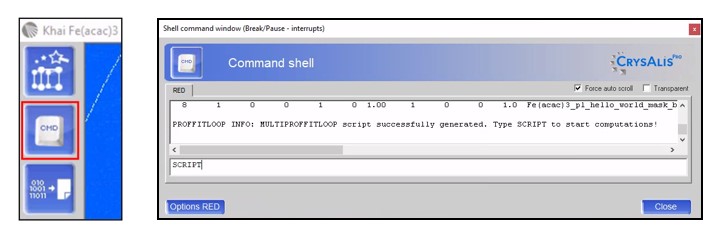
Open a *.par file in the offline version of CrysAlisPro, click on the power tool Command shell, and type “XX PROFFITLOOP” into the command line.

Figure 1. Access the CrysAlisPro feature Proffitloop
After pressing <Enter>, the [Proffit: CrysAlisPro data reduction assistant] wizard will appear, where a user selects default integration settings for all processing jobs. Proceed here as usual. While running Proffitloop, CrysAlisPro uses the same orientation matrix for all processing runs. Once all settings for the six steps are selected, press <Finish>. A new [Proffit loop dialog] GUI will appear, where some parameters can be changed. For more information on these parameters, please visit this link at the Rigaku X-ray Forum.

Figure 2. Proffitloop GUI, which is responsible for the script generation
You can now choose which of the parameters will be used (Single run: Yes), or not be used (Single run: No), or tested for both options (Multi-run: Both) during multiple data reduction. Depending on the different parameter combinations, a number of loops will be generated and saved as *.proffitpars files (see Summary info box. In this case, Number of loops = 8). The files are saved under the following file name template:
- Experiment name (here: “Fe(acac)3”)
- “pl” is an abbreviation for proffitloop and indicates that the respective file was generated by the proffitloop dialog. If you tick the Use short names box, “pl” is removed from the file name.
- “hello_world” can be replaced with additional information given by the user in the edit box on the GUI. Please do not use any special characters. Use lowercase letters. Use underscores (“_”) instead of blank spaces.
- Parameters summary (here: “jp_inc_mask_bkg1_sd”; short names: “j_i_m_b1_s”). Information on which of the parameters were used for the respective data reduction.
Selecting Multi-process proffitloop will increase the processing speed, and consequently decrease the processing time significantly, and thus, is highly recommended. After pressing <OK>, you can now choose the cores for subprocesses. Once you press <Run with sub processes>, please go back to power tool Command shell, type “SCRIPT” into the command line and open multiproffitloop.mac. The Proffitloop script will now run unattended for a few minutes.

Figure 3. How to run the multi-process proffitloop
After successful completion of the script, the files are stored in the experiment folder, and the results can be examined using the power tool Data finalization. The best parameter combination for data reduction for each dataset run can be evaluated by clicking on the power tool icon and scrolling through the data reduction output using the drop-down menu at the bottom of the window. By looking at the statistics printout tables (e.g. I/sigma, Rint values) for each processed dataset, you will be able to identify the best combination of reduction options to produce the best dataset.

Figure 4. Results of each computation can be viewed in the DC RRP window
Note: Threshold-based decisions may change depending on the outcome of the data reduction. The most obvious change that can happen is that different space groups or crystal systems are selected, giving drastically different values and making one result seem potentially worse than it is in reality.
Author

Rigaku Europe SE | Germany
Dr. Khai-Nghi Truong obtained his PhD in inorganic chemistry and small molecule crystallography at RWTH Aachen University (Germany) in late 2018, working under Prof. Ulli Englert on the synthesis and characterization of metal-organic frameworks. From 2019 to 2022, Khai worked as a postdoctoral research fellow with the renowned Prof. Kari Rissanen at the University of Jyväskylä (Finland). He was part of the EU funded Horizon2020 FET Open research and innovation programme INITIO working as an expert in solid-state. His areas of expertise are inorganic and organic small molecule crystallography, supramolecular chemistry as well as materials science. As service crystallographer, chair of the Young Crystallographers of the German Society of Crystallographic (DGK-YC) and German representative of the Young Crystallographers’ European Crystallographic Association (YC-ECA) for several years, Khai has build-up a strong network of collaborators all around the world. He has contributed to more than 60 publications using different diffraction methods, viz. X-ray powder diffraction, single crystal X-ray and neutron diffraction. Khai joined Rigaku Europe SE in mid 2022, working as Application Scientist for both single crystal X-ray and electron diffraction supporting Rigaku’s clients in the EMEA region. Want to learn more? Connect with Khai-Nghi Truong, PhD LinkedIn .

Subscribe to the Crystallography Times newsletter
Stay up to date with single crystal analysis news and upcoming events, learn about researchers in the field, new techniques and products, and explore helpful tips.

Contact Us
Whether you're interested in getting a quote, want a demo, need technical support, or simply have a question, we're here to help.