The Molecular Grabber Method: Another Crystalline Sponge using Protein
Authors
Takashi Matsumoto¹, Akihito Yamano¹, Ryosuke Nakashima², Kunihiko Nishino²
¹Rigaku Corporation – 3-9-12 Matsubara-cho, Akishima-shi, Tokyo 196-8666, Japan, ²Institute of Scientific and Industrial Research, Osaka University, 8-1 Mihogaoka, Ibaraki, Osaka 567-0047, Japan
Abstract
Molecular structure determination is beneficial for the development of medicines, aroma chemicals, and agrochemicals. Single crystal X-ray diffraction (SC-XRD) analysis is the most powerful technique for molecular structure determination. However, SC-XRD analysis requires good quality crystals.
In fact, the biggest hurdle for SC-XRD analysis is crystallization. Crystallization trials require a large amount of highly purified target compounds. Moreover, good quality crystals for SC-XRD analysis might not be obtained despite performing tedious and time-consuming trials. In this case, we have to abandon the direct structure determination by SC-XRD. As one way to address this situation, Fujita et al. have reported the crystalline sponge method (CS method) for the structure determination of small molecules [1]. However, as with other analysis techniques, the CS method has some limitations.
The CS method utilizes metal-organic frameworks (MOFs) as the pre-crystallized ‘‘container” for the analytes. The “container” is equipped with flexible features to fit various analytes and must have enough space to accommodate a wide variety of molecules. An MOF is a large structural object with three-dimensional networks; thus, the spaces to accommodate molecules have limitations in principle.
To overcome the above difficulty, we came up with the idea of a “molecular grabber,” utilizing an easy-to-crystallize protein that has a multi-site binding pocket to bind a variety of types of molecules and gives high-resolution spots.
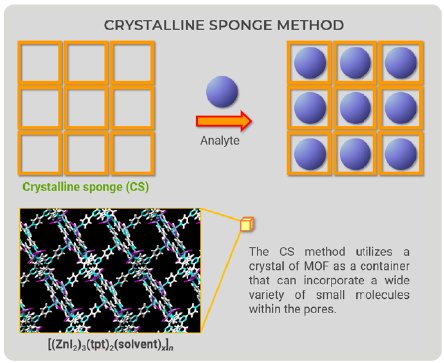
Figure 1: Overview drawing of the CS method.
Selection of protein as “container”
-transcriptional repressor-
The CS method utilizes a crystal of MOF as a container that can incorporate a wide variety of small molecules within the pores. Looking at CS method figures (Fig. 1), we came up with a new idea—the container would not have to be an MOF (Fig. 2). We could prepare a “container” with the ability to bind to a wide variety of compounds; therefore, we focused on proteins because they can bind to organic compounds.
We thought that using proteins as the “container” had five requirements: the ability to bind to a wide variety of compounds, stability, a simple purification procedure, easy to crystallize and obtaining good quality crystals. We searched for a protein that fulfilled these requirements and found that RamR, a transcriptional repressor, was suitable for the container [2]. RamR controls the expression of the multidrug efflux transporter AcrB gene and can bind to a wide variety of compounds. Moreover, RamR is a stable protein and is easily purified
and crystallized and produces crystals with high-resolution spots. For these reasons, we decided to explore the possibility of using RamR as the container to determine the structures of unknown compounds instead of using MOFs.
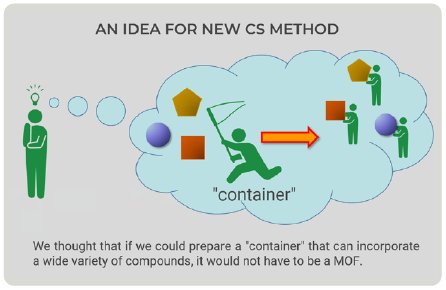
Figure 2: New idea for new CS method.
Experiments
In the CS method, the compounds are taken up by soaking but in this method RamR-compound complexes were prepared by co-crystallization. RamR and the compound solution were mixed and incubated overnight. Crystallization was carried out by the sitting drop vapor diffusion method at 25ºC (Fig. 3) and, after 1 week, block-shaped crystals appeared. All diffraction images were collected on a Rigaku XtaLAB Synergy Custom diffractometer (Cu Kα) at 100 K. The diffraction data were processed using CrysAlisPro [3]. The structure of each complex was solved by molecular replacement, which is an easy phase determination method.
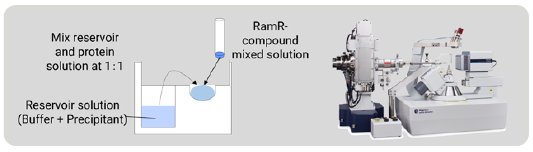
Figure 3: Co-crystallization of the RamR-compound com-plex and the diffractometer for the SC-XRD measurement.
Results and discussion
First, we tried to analyze ethidium, which is familiar in the field of molecular biology and is used as a staining agent for nucleic acids. After structure analysis, the electron density map of the grabbed molecule was clearly visible. From the shape of the density map, ethidium is easily identified (Fig. 4). The ethidium molecule fits into the binding pocket of RamR and binds to amino acids in the binding pocket via hydrogen bonds and π-π stacking interactions.
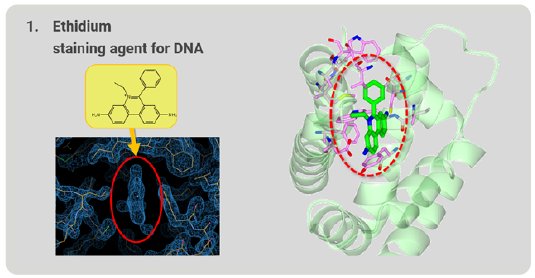
Figure 4: Ethidium grabbed by RamR.
Next, we investigated whether RamR can bind large, rigid cholic acid, which is too large and bulky to be accommodated in the binding pocket. Cholic acid has a steroid skeleton. After structure analysis, the electron density map of the grabbed molecule was clearly appeared. From the shape of the density map, cholic acid is easily identified (Fig. 5). RamR bound the cholic acid using different amino acid residues compared with ethidium. This complex used hydrogen bonds and hydrophobic interactions without π-π stacking interactions.
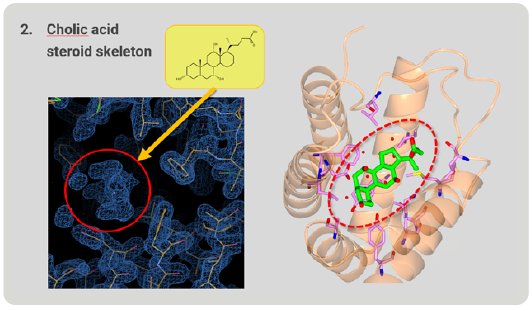
Figure 5: Cholic acid, a large molecule grabbed by RamR.
Figure 6 is a superimposition of RamR-ethidium complex (light green) on RamR-cholic acid complex (orange). Surprisingly, the structure of the binding pocket of the RamR-cholic acid complex was quite different from that of the RamR-ethidium complex. The α7b and α8a helices of the RamR-cholic acid complex, indicated by the arrows, were completely uncoiled. Furthermore, the α7a helix and uncoiled α7b shifted outward, resulting in an expansion of the volume of the binding pocket to accommodate the bulky rigid cholic acid molecule. This flexibility is another feature of RamR.
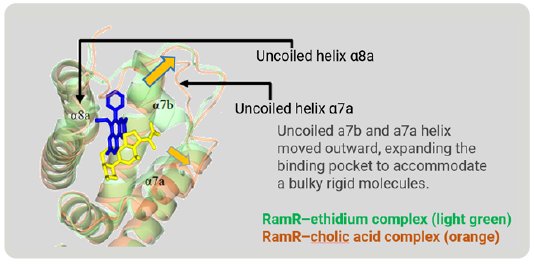
Figure 6: Superimposed image of RamR–ethidium com-plex (light green) on RamR–cholic acid complex (orange).
Dequalinium has a long, flexible carbon chain that is too long to be accommodated in the binding pocket. The α6, α7b, α8a, α8b, and α9 helices shift outward to expand and reshape the binding pocket to accommodate this long, flexible compound (Fig. 7). In this way, RamR can grab organic compounds by flexibly changing the shape of the binding pocket and optimizing it to the shape of the target compound. Because of this feature, we named this method the “Molecular Grabber” method.
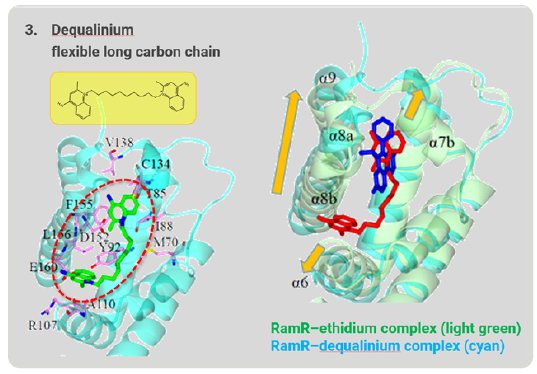
Figure 7: Long-chain compound grabbed by RamR.
We investigated whether this method could be used to determine the structures of unknown compounds. Gefitinib intermediate 1 is a synthetic intermediate of gefitinib, a well-known anti-cancer medicine. As a result of structure analysis, a good electron density map appeared and, from this shape, it was revealed that gefitinib intermediate 1 has a thin and flat structure (Fig. 8). As a result, it was proved that Molecular Grabber method can reveal the structure of unknown compound.
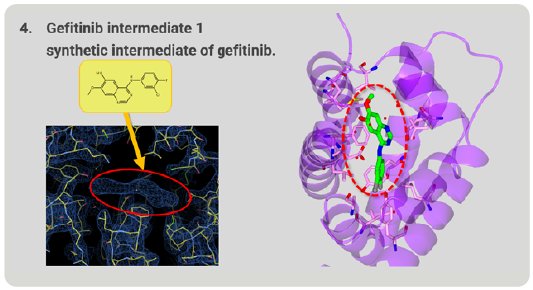
Figure 8. Structure of unknown gefitinib intermediate 1 grabbed by RamR.
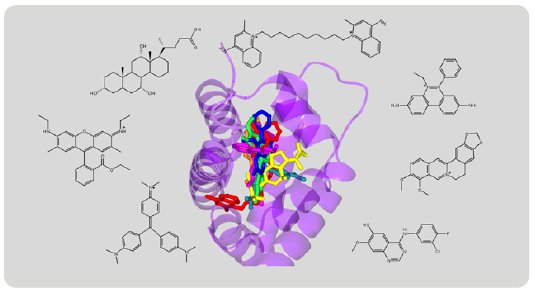
Figure 9: Superimposition of various compounds grabbed by RamR.
Summary
The structures of various compounds have been revealed by the Molecular Grabber method (Fig. 9). Like a hand grabbing a lemon or a string, RamR grabs a wide variety of compounds and reveals their structures. Based on these structures, RamR flexibly changes the volume and shape of its binding pocket to match the size and shape of target compounds and can then grab the compound. Like the CS method, the molecular grabber method has the potential to be used for X-ray structural analysis of organic compounds that are difficult to crystallize.
t is also important to note that RamR is a water-soluble protein. RamR prefers water and is tolerant to alcohol and DMSO. Therefore, this method can be used to work on compounds that are dissolved in polar solvents. We hope that the molecular grabber method will contribute to developing innovative new drugs, aroma compounds, and agrochemicals and identifying residual intermediates. This method does not require any license to use, so it is also a significant feature that any interested researcher can try and use.
References
[1] Inokuma, Y., Yoshioka, S., Ariyoshi, J., Arita, T., Hirota, Y., Takada, K., Matsunaga, S., Rissanen, K. & Fujita, M. (2013). Nature 495, 461-467.
[2] Matsumoto, T., Nakashima, R., Yamano, A. & Nishino, K. (2019). Biochem. Biophys. Res. Commun. 518, 402-408.
[3] Matsumoto, T., Yamano, A., Sato, T., Ferrara, J. D., White, F. J., Meyer, M. (2021) J. Chem. Crystallogr., 51, 438–450.

Contact Us
Whether you're interested in getting a quote, want a demo, need technical support, or simply have a question, we're here to help.
