Application of the Crystalline Sponge Method for Structure Determination of Persistent organic Pollutants of Agrochemical Degradation Products
Abstract
Identification of agrochemical degradation metabolites occurring in soil, water, and crops, and assessment of their toxicity, are of great importance in view of food safety. Single crystal X-ray (SCX) analysis, the most powerful tool to determine a three-dimensional molecular structure, has rarely been utilized for agrochemical metabolites because only a trace amount of them can be isolated from the environment and crops, which makes it very difficult to obtain a suitable single crystal for SCX analysis. The crystalline sponge (CS) method is a novel technique for SCX sample preparation developed by Prof. Fujita,¹ which utilizes a crystal of a metal-organic framework (MOF) as a pre-crystallized molecular container that can incorporate a wide variety of small molecules within the pores and arrange the molecules in a regular pattern according to the periodicity of the host crystal. SCX analysis of the resultant inclusion crystal facilitates direct observation of the molecular structure of the target analyte. Here we report the initial examples of the structure identification of agrochemical metabolites from only a few micrograms of them. We demonstrate that the SCX analysis clearly distinguished the metabolites with very similar structures, which is extremely challenging for a conventional method using nuclear magnetic resonance (NMR) and mass spectrometric (MS) analysis.
Introduction
For future production and use of agrochemicals based on green, sustainable chemistry, there are urgent demands for identifying agrochemical degradation metabolites occurring in soil, water, the atmospheric environment and crops, and for assessing their toxicities in view of food safety. In the determination of the chemical structures of such unidentified metabolites, a common approach is to synthesize these as reference standards based on the structures estimated from chemical information obtained from mass spectrometric analysis, by precision mass spectrometric analysis using Liquid Chromatography / High-Resolution Mass Spectrometry (LC/HRMS), GC/HRMS and nuclear magnetic resonance (NMR) spectral analysis, and to identify unknown metabolites by comparing with the chemical identities of the reference standards.² However, agrochemical degradation metabolites are usually obtained in only very small amounts, and a multitude of structures can occur in the environment and in crops. Therefore, synthesizing all such potential metabolites and similar candidate compounds is not a realistic endeavor due to the extreme time and economic efforts involved.
Crystalline sponge method
In conventional crystallization, the process predominantly depends on physical and chemical properties, which makes it challenging or even impossible to obtain a single crystal of a target compound suitable for SCX analysis (Figure 1a). In addition, a relatively large sample amount (milligram scale) is required for conventional crystallization trials. The crystalline sponge (CS) method developed by Fujita’s group overcomes these drawbacks of conventional crystallography.1 In the CS method, a single crystal of a metal organic framework (MOF) is utilized as a pre-crystallized molecular container (Figure 1b). When the CS crystal is soaked in a solution of analyte, the analyte molecule is incorporated and ordered within the pores of the CS as a consequence of molecular recognition by the CS framework. After the soaking process takes place, without losing the crystallinity of the CS, SCX affords a reliable molecular structure of the incorporated analyte. The sample amount required for the soaking is only few micrograms, sufficient to fill the CS pores (Figure 1c).
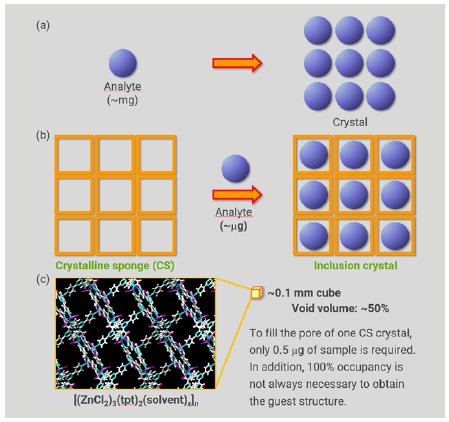
Figure 1: Comparison of conventional crystallization and the CS method.
Experiments
We chose five compounds of agrochemicals and their expected metabolites as analytes, including pentachlorophenol (PCP), its metabolites, 2,3,4,5-tetrachlorophenol (TCP) and 2,3,5,6-TCP, and metabolites of 1,1-bis(4-chlorophenyl)-2,2,2-trichloroethane (DDT), 4,4’-dichlorobenzophenone (4,4’-DBP) and 4,4’-dichloro-2,2-diphenylethanol (4,4’-DDOH) (Figure 2a). PCP and DDT were eliminated by Stockholm Convention on Persistent Organic Pollutants (POPs) in 2001, and their residue and degradation in environment and crops has attracted much attention.
Preparation of an inclusion crystal was implemented using reported methods (Figure 2b).1,3 To one single crystal of CS stored in 50 µL of cyclohexane in a glass vial, a dichloromethane solution of an analyte (1-5 µg) was added. The vial was sealed with a plastic cap pierced with a syringe needle and was kept at room temperature to allow the solvent to be evaporated slowly to dryness. The resultant inclusion crystal was picked up and mounted on Rigaku XtaLAB Synergy-S or XtaLAB Synergy-Custom FR-X diffractometer (Cu Kα) to obtain diffraction images. Data collections were conducted at 100 K, and were typically completed in 1–12 hours with 100% completeness in a resolution range of > 0.83 Å.
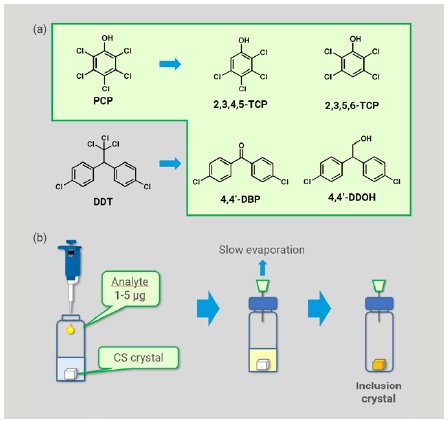
Figure 2: (a) POPs agrochemicals and their expected metabolites investigated and (b) typical sample preparation procedure.
Results and discussion
First, we investigated PCP. A 1 µg solution of PCP was added to one particle of CS to prepare an inclusion crystal (Figure 3a). SCX analysis of the inclusion crystal showed PCP molecules firmly trapped in the pores of the CS to form a columnar stacking structure with π-π interactions between the guest molecules (Figure 3b). Selective display of the guest molecule alone clearly showed the PCP structure (Figure 3c). Taking advantage of the SCX analysis, we were able to clearly distinguish between the substituent oxygen atoms and chlorine atoms in the PCP based on the difference in the magnitude of electron density (Figure 3d). We succeeded in the direct determination of the structure of PCP using an extremely small sample amount totaling 1 µg.
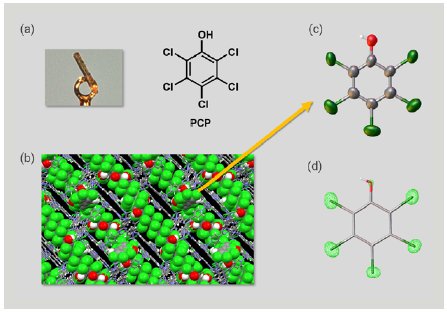
Figure 3: (a) Inclusion crystal of PCP, (b) packing structure of the inclusion crystal, (c) ORTEP drawing of PCP, and (d) electron density map of PCP.
Next, two isomers of TCP were investigated. A 5 µg solution of 2,3,4,5-TCP or 2,3,5,6-TCP was added to one particle of each CS to prepare inclusion crystals. As with PCP, SCX analysis of the inclusion crystals confirmed the molecular structures of the TCPs (Figure 4a, b). Notably, the chlorine atom substitution sites in the two isomers were clearly able to be distinguished based on the electron density maps (Figure 4c, d). Because the two isomers have identical elemental composition and few ¹H nuclei, it is difficult to distinguish them by NMR or MS. As suggested by this case, degradation products resulting from the same substrate often have a similar structure, so SCX represents a powerful tool for degradation product analysis.
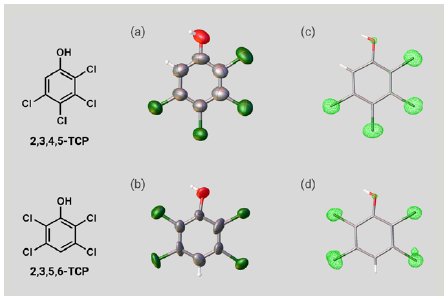
Figure 4: ORTEP drawings of (a) 2,3,4,5-TCP and (b) 2,3,5,6-TCP. Electron density maps of (c) 2,3,4,5-TCP and (d) 2,3,5,6-TCP.
We also investigated two expected metabolites of DDT. A 5 µg solution of 4,4’-DBP or 4,4’-DDOH was added to one particle of each CS to prepare inclusion crystals. SCX analysis of the inclusion crystals clearly confirmed the molecular structures of 4,4’-DBP and 4,4’-DDOH (Figure 5a, b). Although the molecular shapes of the expected DDT metabolites are significantly different from those of the PCP metabolites, the large and flexible CS pores efficiently incorporated them. These results show that the CS method can cover a wide variety of molecular structures and is applicable to metabolites of various POPs agrochemicals.
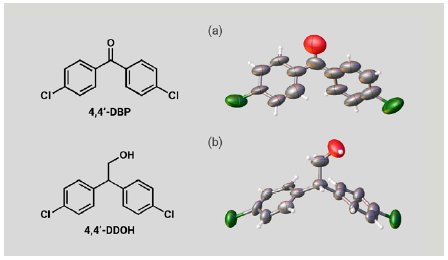
Figure 5: ORTEP drawings of (a) 4,4’-DBP and (b) 4,4’-DDOH.
Summary
We demonstrated that the CS method can be a powerful tool for structural determination of assumed POPs agrochemical degradation metabolites using extremely small sample amounts. As no other approach can produce accurate structural determinations with just micrograms of unidentified sample purified by preparative HPLC, based on degradation /metabolism experiments on a laboratory scale, SCX combined with the CS method thus offers the potential of dramatically changing the conventional modality of degradation metabolite analysis. This achievement will lead to the development of new agrochemicals with higher environmental and crop safety and could allow the elucidation of detailed information on the degradation metabolism pathways of residual organic contaminants, such as existing agrochemicals and POPs, which have not been clarified so far.
References
[1] Y. Inokuma, S. Yoshioka, J. Ariyoshi, T. Arai, Y. Hitora, K. Takada, S. Matsunaga, K. Rissanen and M. Fujita: Nature 495, 461–466 (2013).
[2] K. Takagi: J. Pestic. Sci. 45, 119–123 (2020).
[3] Y. Inokuma, S. Yoshioka, J. Ariyoshi, T. Arai and M. Fujita: Nat. Protoc. 9, 246–252 (2014).

Contact Us
Whether you're interested in getting a quote, want a demo, need technical support, or simply have a question, we're here to help.
